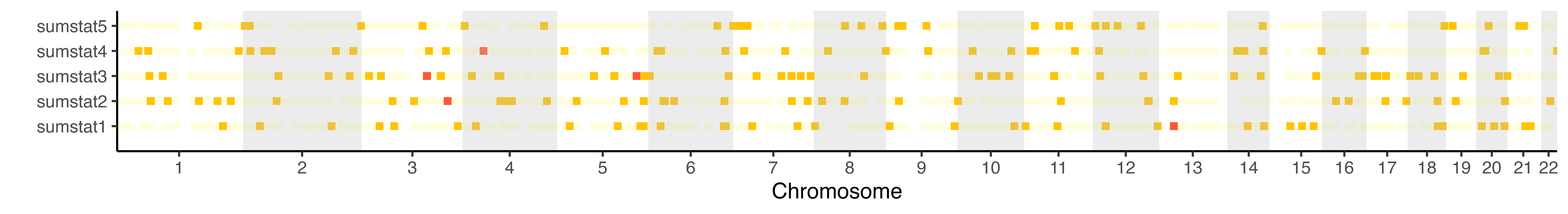
The goal of ggmugs is to visualize multiple Genome-wide Association Study (GWAS) summary statistics
To install the ggmugs package, run:
if(!require("devtools")){
install.packages("devtools")
library("devtools")
}
devtools::install_github("Broccolito/ggmugs")This is a basic example which shows you how to solve a common problem:
library(ggmugs)
plt = ggmugs(
study_name = c("sumstat1", "sumstat2", "sumstat3", "sumstat4", "sumstat5"),
summary_stat = c("https://raw.githubusercontent.com/Broccolito/ggmugs_data/main/sumstat1.txt",
"https://raw.githubusercontent.com/Broccolito/ggmugs_data/main/sumstat2.txt",
"https://raw.githubusercontent.com/Broccolito/ggmugs_data/main/sumstat3.txt",
"https://raw.githubusercontent.com/Broccolito/ggmugs_data/main/sumstat4.txt",
"https://raw.githubusercontent.com/Broccolito/ggmugs_data/main/sumstat5.txt"),
p1 = 1e-4,
p2 = 1e-6,
p3 = 1e-8,
color1 = "#FFFFE0",
color2 = "#FFC300",
color3 = "#FF5733"
)Here is an example output from the function: